A survey of ancient paralog expression patterns in Arabidopsis thaliana root cell-types
Published:
I’ve decided it would be nice, for academic posterity, to include a sample of my work on ancient gene duplicates in Arabidopsis thaliana. I’ve quoted the abstract below but fair warning: it’s rife with evo-genetic neologisms.
Whole-genome duplication events have played an extensive role in the evolution of flowering plants. The sudden doubling of genetic material can expedite large scale changes in gene function and expression patterns. This process can be an adaptively beneficial source of genetic variation.
In the context of paleopolyploids, wherein there are identifiable remnants of an ancestral polyploidy event in a diploid species, this often facilitates the diversification of gene families as these duplicated genes are no longer selectively constrained to one specific function or regulatory pattern. Alternative splicing (AS) offers an avenue with which genes duplicated by these polyploidy events may further contribute to the functional complexity of the cell.
While the extent to which AS influences functional or regulatory divergence in ancient paralogs has been previously investigated, such as those derived from the α-WGD event in Arabidopsis thaliana, the manifestation of this divergence in distinct tissues and cell-types is less understood. Using RNA-Seq data I have surveyed the transcriptomes of root developmental zones and their constituent cell-types in paleopolyploid Arabidopsis thaliana. Incorporating such high-resolution data with further in silico analyses, this demonstrates near-complete divergence in both gene expression and splicing patterns.
In other words, I wanted to see how gene expression between ancient duplicates changes over time. For instance, how many genes show conserved expression patterns following several million years of evolution? Additionally, is there evidence of alternative splicing event conservation between these duplicates? Moreover, I wanted to see how many - if any - were shared between different cell-types in the Arabidopsis root.
To do this I was interested into breaking conservation down into two categories (inspired by Tack et al. 2014):
(1) Symmetric conservation: whether a given pair of duplicate genes or splicing events mutually expressed (i.e. expression symmetry)
(2) Qunatitative conservation: whether a given (symmetric) duplicate pair was differentially expressed.
In doing so there were two major aspects of the analysis. Below I’ve highlighted a quaint teaser of the results.
Methods
As a brief digression, I’d like to outline some important aspects of the methodology. The figure below describes the difference between symmetric conservation (i.e. qualitative) and quantitative conservation (left and right subplots, respectively) for both gene expression and alternative splicing (top and bottom).
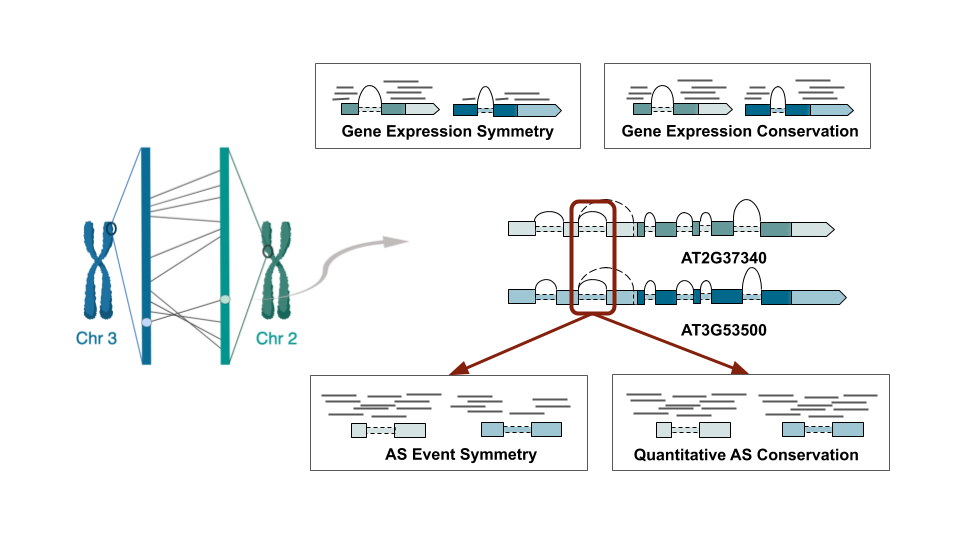
As you can see, symmetry simply depends on whether a duplicate pair is expressed at the same time in the same cell. Otherwise, I referred to these as divergent (or asymmetric).
Exploring quantitative differences was a bit more convoluted. To explore differential gene expression, I used edgeR - a commonly used R package which performs analyses of differential gene expression (DGE). Unfortunately, quantitative comparisons from within the same genome introduce a suite of problems, most notably: (1) paralogs may be of different in length (meaning longer genes will be biased towards more sequencing reads, obscuring reliable comparison) and (2) these are intra-sample comparisons so they share the same library size. To do this, I used a stripped-down version of the edgeR protocol that introduced matrix offsets that incoporoated gene lengths and sample-specific library sizes in hopes to reduce the likelihood of false-positives.
For alternative splicing I used a modified of the python-based AS detection pipeline described in Chalhoub et al. 2014 and Tack et al. 2014 to identify the possible AS events. To assess whether AS events diverged quantitatively I modified one of my R scripts to explore differential alternative splicing based on fitting to the following logistic regression model:
pr(alt)∼bi(p=logit-1 (b0+bcondition))
In this case, pr(alt) is proportion of alternatively mapped reads compared to all splice-junction spanning reads. The purpose was to consider at the proportion of alernatively mapped reads as a “hit” or “success”, compared to “failures” - i.e. constitutively mapped reads.
Cell-type specific splicing conservation
With all that said, one my first analyses was to use both gene expression and alternative splicing data to assess how duplicates across 15 root cell types have diverged:

Interestingly, gene expression and alternative splicing differ substantially in their patterns of conservation. Many genes tend to expressed symmetrically in all fifteen cell types; however, they are frequently expressed at significantly different levels, with only 10 duplicate pairs exhibiting pan-root conservation.
In contrast, most alternative splicing events are not symmetrically expressed, and when they are, they tend to be found in only a few cells types (i.e. both 14/15 to 11/15 cell types do not express either events). These events are somewhat often quantitatively conserved, but this occurs rarely (22 out of 1871 events pairs) compared to all splicing events in a given cell.
Conclusion: Overall, it appears most paralogs differ considerably in their regulation.
Characteristics of paralogous AS events
Evidently there are few splicing events that exhibit perfect conservation - in terms of expression symmetry or splicing ratios - but what about partially conserved events? Is there any information that be gleaned from looking at symmetry a characteristic of individual gene pairs?
To accomplish this I assigned a symmetry percentage to each to gene pair based off their shared number of alternative splicing events compared to asymmetric events.
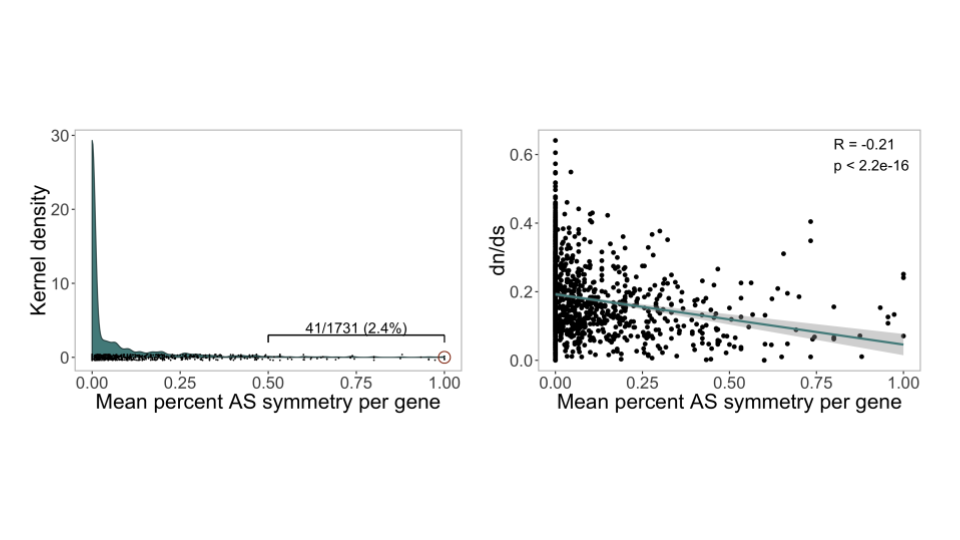
Looking at the mean symmetry across all cell types, it was clear that there was still very little conservation.
I was then curious about what might be driving the conservation of these scant few events. As you can see in the figure above, I performed a series of dn/ds analyses between the CDS of duplicate gene pairs and used that as a proxy for the rates of sequence divergence. Interestingly, although the correlation is somewhat weak, genes that tended to exhibit higher levels of symmetry were also less susceptible to sequence change.
It is possible these genes are important! To assess whether they might share interesting (or consistent) function properties, I ran a gene-set enrichment analysis (GSEA) on partially conserved gene pairs (>25% events conserved) against all similarly aged gene duplicates.
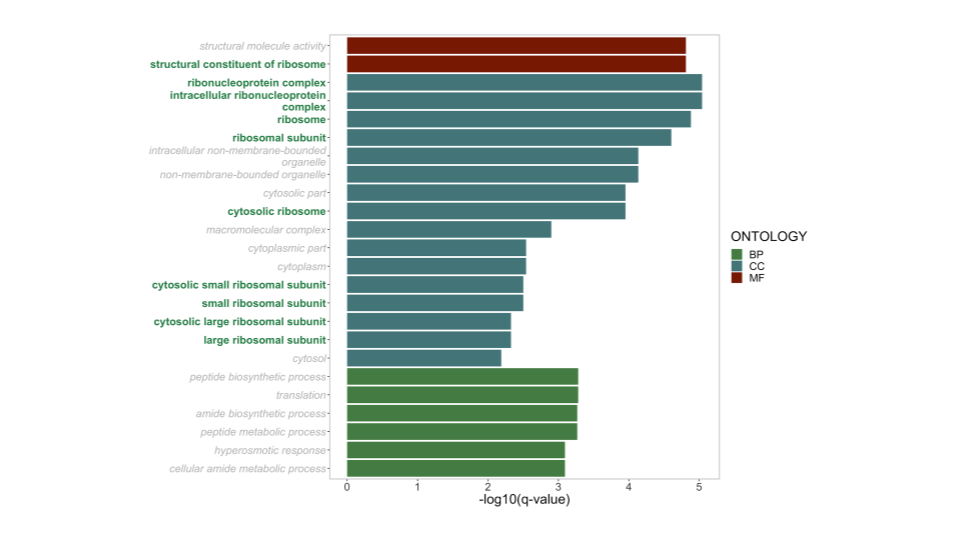
As this material is yet to be published, I’m reticent to go into much more detail. However, it appears ribosome-associated gene duplicates display higher retention of alternative splicing events!
References
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. science. 2014 Aug 22;345(6199):950-3.
Tack DC, Pitchers WR, Adams KL. Transcriptome analysis indicates considerable divergence in alternative splicing between duplicated genes in Arabidopsis thaliana. Genetics. 2014 Dec 1;198(4):1473-81.
