Analysis of multiple Brassica transcriptomes reveals subgenome dominance in the response of Brassica napus to Sclerotinia sclerotiorum
Published:
This was perhaps my favourite project in graduate school. I was interested in seeing how polyploids responded to biotic stress - specifically subgenome-specific differences in stress response.
For a bit of background, this is an exerpt of my abstract:
Whole-genome duplication (WGD; polyploidy) events have played an extensive role in the evolution of flowering plants. The sudden doubling of genetic material can expedite rapid novel changes to polyploid transcriptomes. For example, polyploids formed via an interspecific hybridization of closely related species, known as allopolyploids, can exhibit inconsistent expression patterns between their parental genome. These incipient disparities in parental gene dosage can have profound effects on the transcriptome of newly formed polyploids, which in turn can influence their response to environmental stressors. In particular, the extent to which the transcriptomic shock of polyploidization modulates the biotic stress response of plant species remains a nascent topic in polyploidy research.
To do this, I grew four different Brassica species: B. napus, its progenitor species B. oleracea and *B. rapa, and a resynthesized B. napus (formed directly from the parental genotypes used in the experiment). These species were then subject to pathogen treatments (both mock-inoculations and fungal-inoculations) from the necrotroph Sclerotinia sclerotiorum. Using three biological replicates per condition, this totalled in 24 RNA libraries which were run on the Illumina NovaSeq 6000 platform.
Now, as much as I’d like to dive into the details, findings, and methodology of the computational aspects of the project, the manuscript for this project yet to be published so I’d prefer to keep things brief.
An interesting side-story that arose from the main project involved the relationship between alternative splicing and gene expression.
First, I performed a cursory set of DGEAs on Brassica hosts using a stringent cutoff of abs(log2FC) > 3, revealing that roughly 10% of polyploid genes and 7% of the diploid genes were highly responsive to the pathogen.
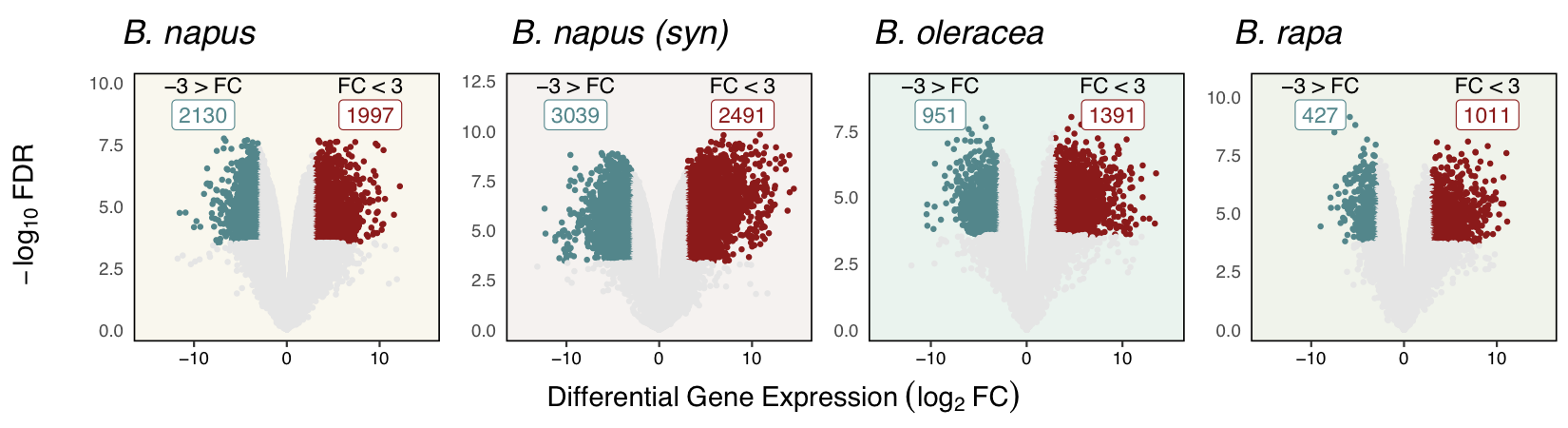
As you can see, there was widespread transcriptome modifications as a result of S. sclerotiorum infection across all species. Now, to identify possible GO term enrichment (and to perform inter-specific GO comparisons) all host genes were assigned GO terms using the same interproscan-based annotation pipeline.
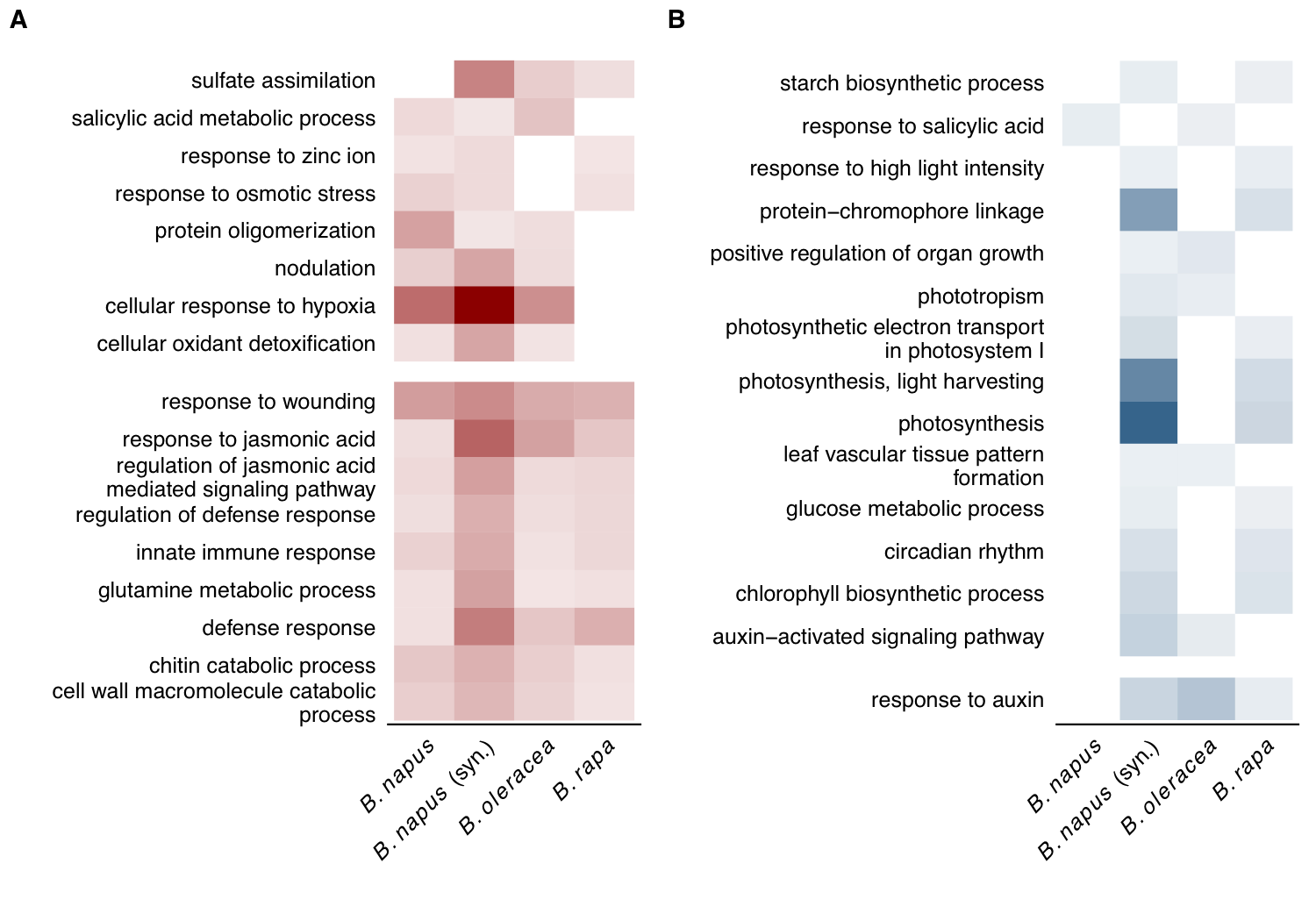
As one would expect based on earlier research involving the S. sclerotinia - B. napus pathosystem, up-regulated genes show significant enrichment of genes implicated in pathogen-associated molecular pattern (PAMPs) and damage-associated molecular pattern (DAMPs) responses - e.g. response wounding, defense response, jasmonic acid associated terms, etc.
Alternative visualization based on average response of particular pathways:
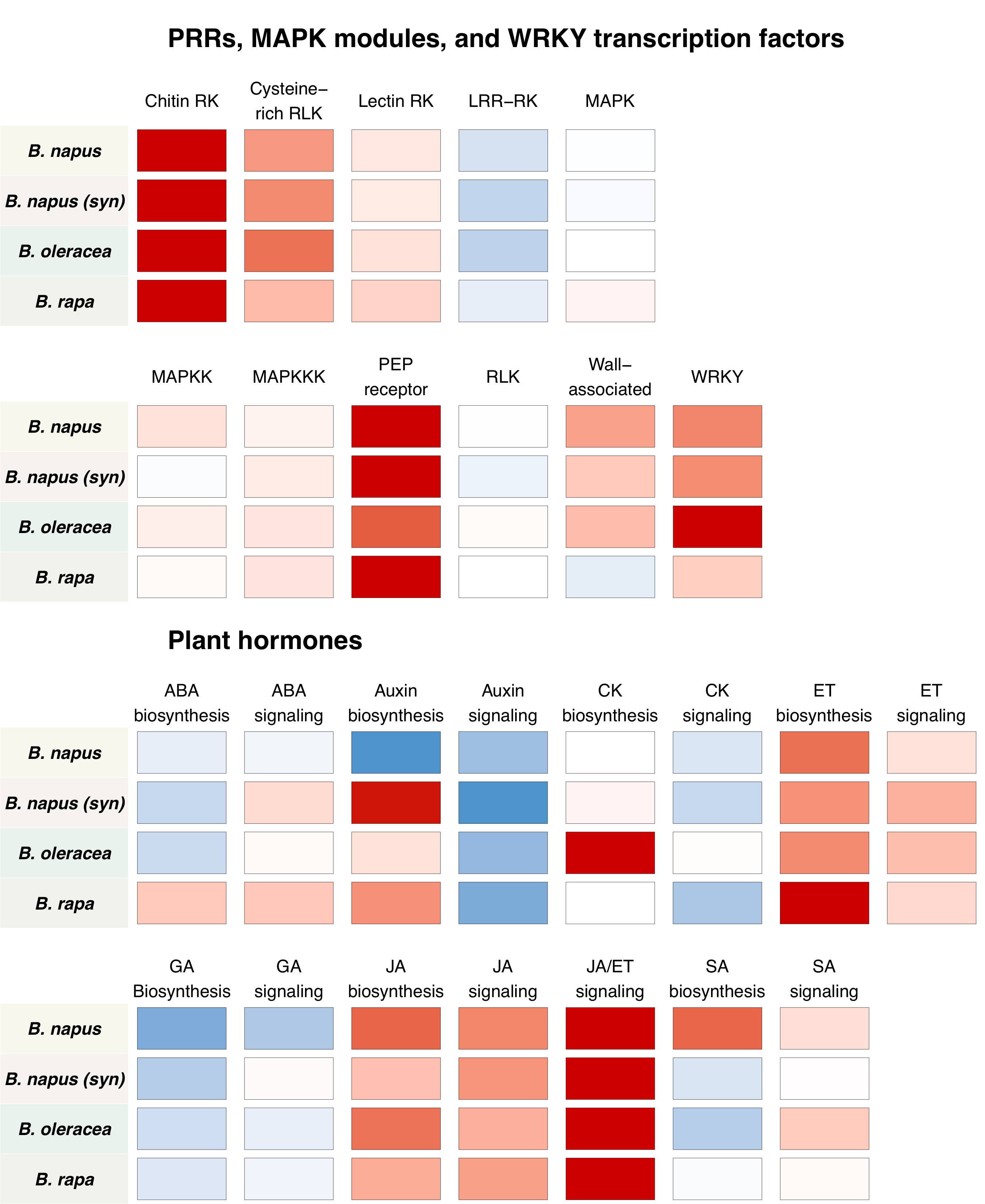
Furthermore, down-regulated genes were significantly enriched for energy metabolism and photosynthesis, suggesting resource-shuffling from constutitve biological processes to accomodate plant innate immune response. This might seem expected, but its especially relevant in the context of alternative splicing.
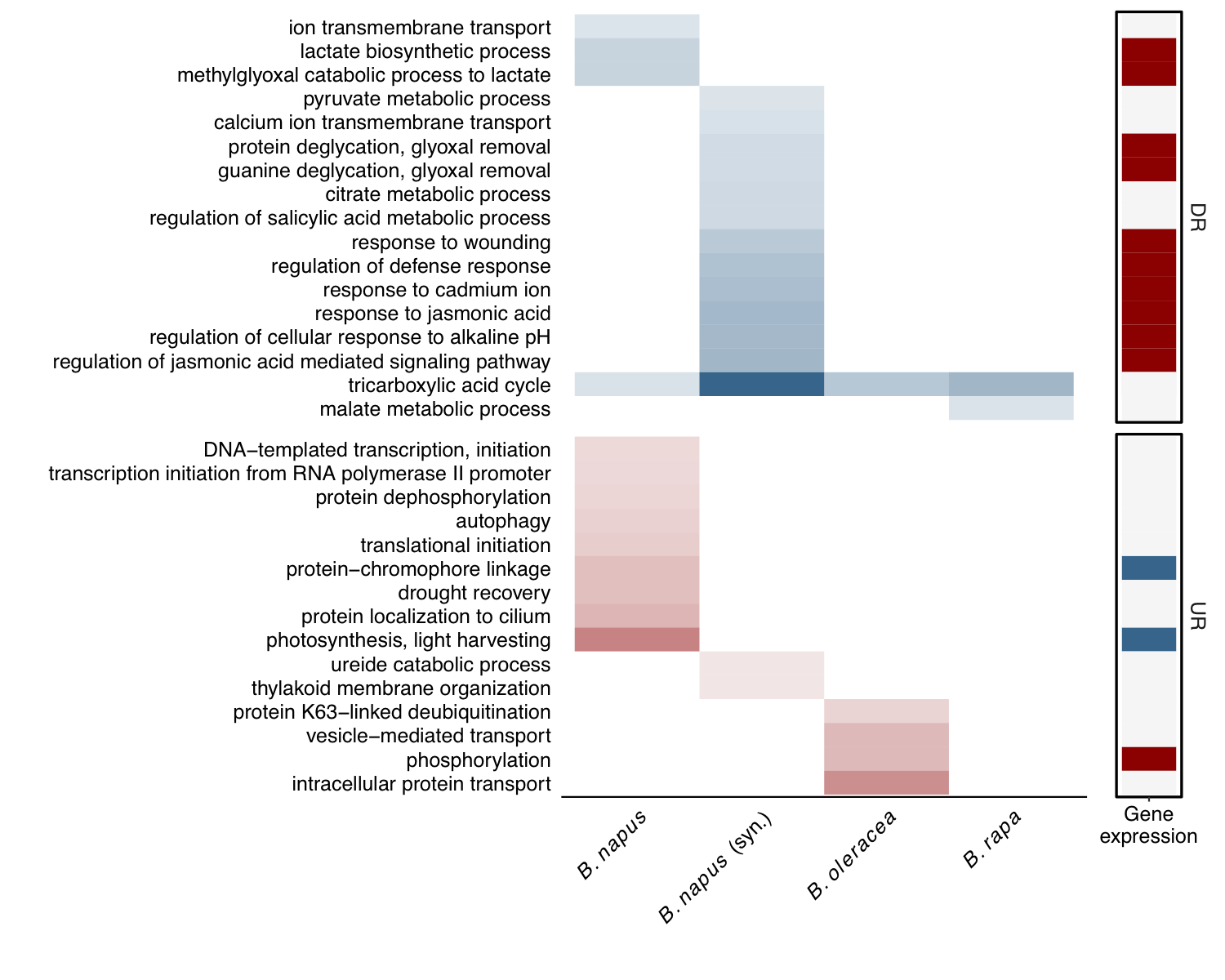
Interestingly, when these same analyses are performed on differentially spliced genes, an opposite trend emerges. Biotic stress and immune response genes experience highly attenuated alternative splicing.
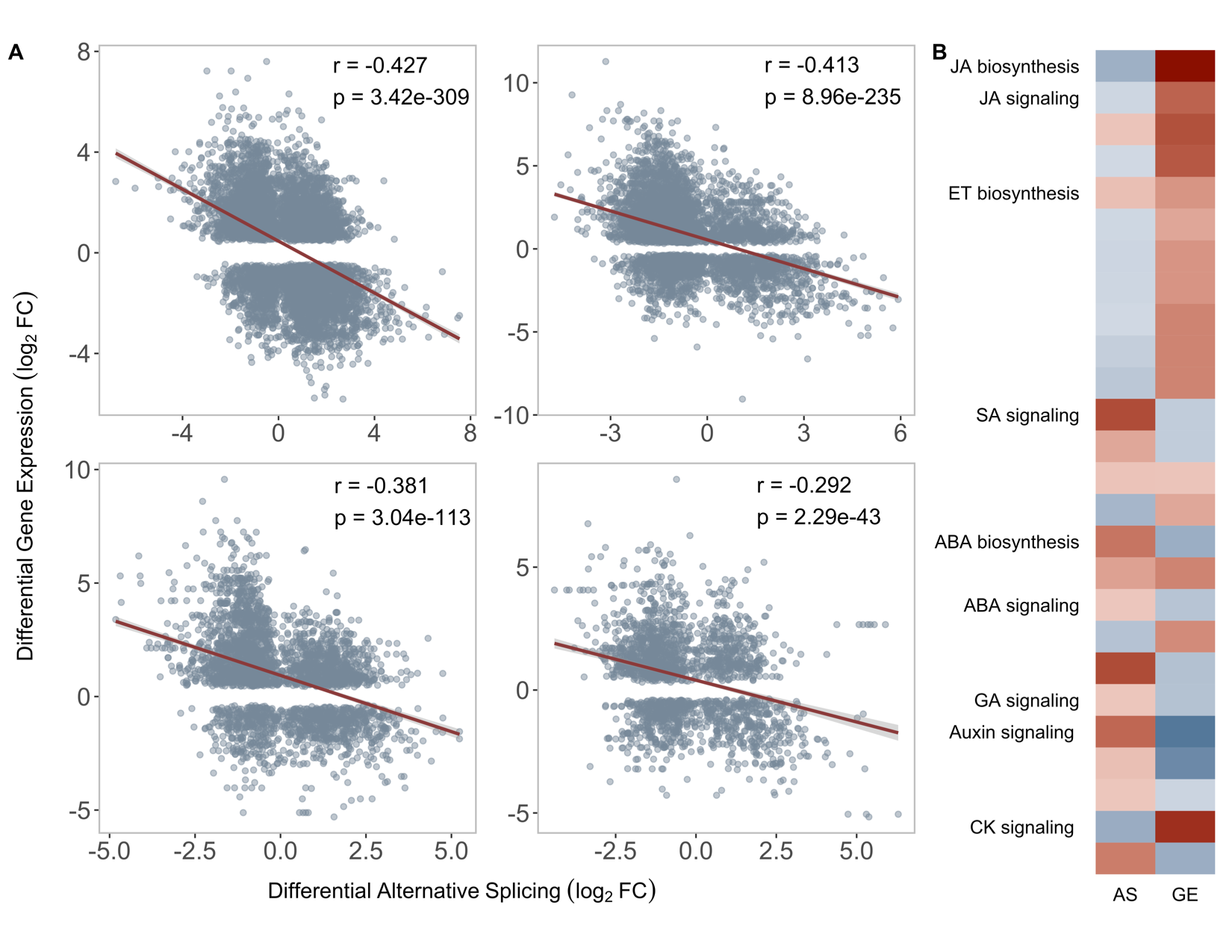
Looking at each host independently, there does appear to be somewhat of a negative correlation, suggesting that many biotic stress genes experience highly reduced splicing under stress.
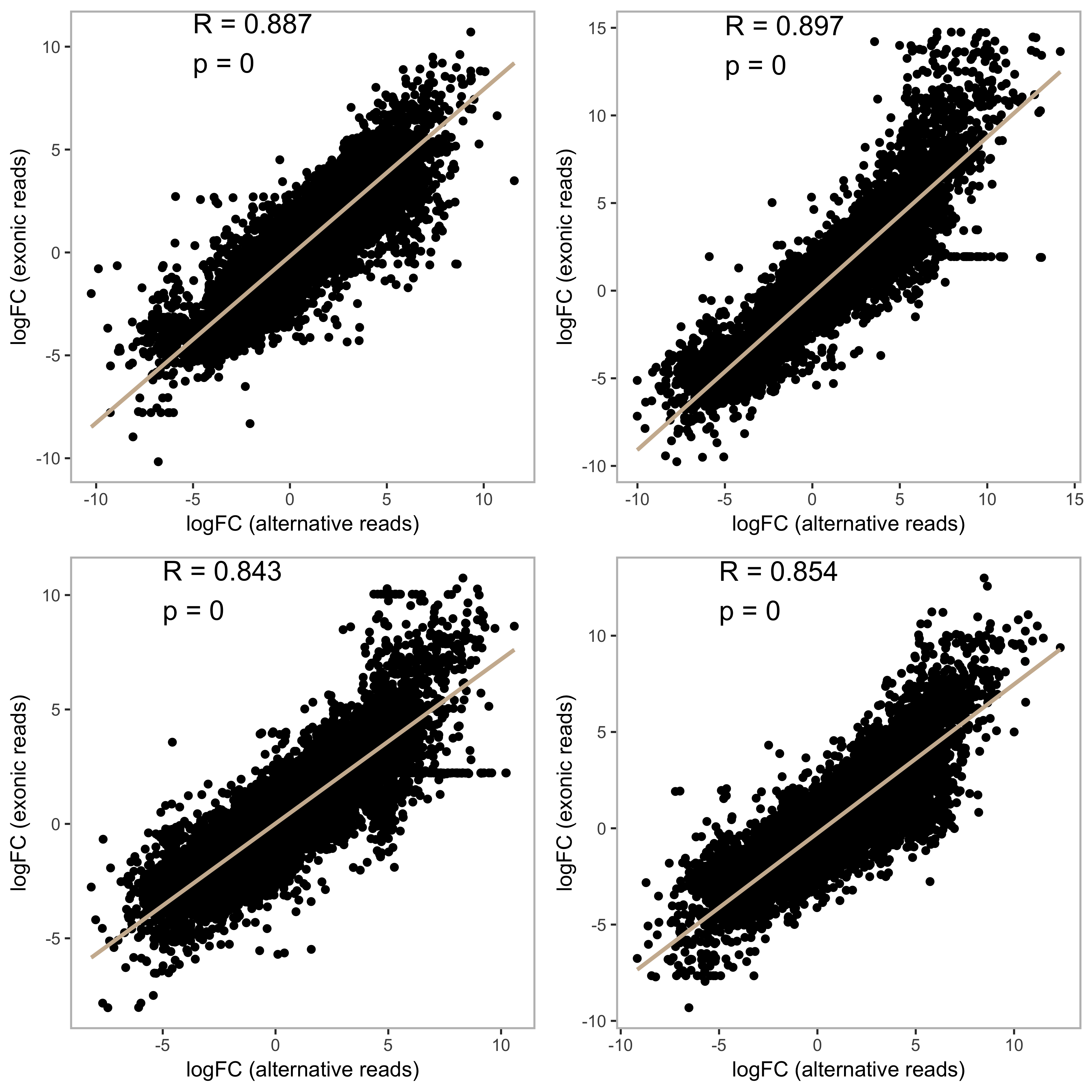
Of course, I thought this could be an issue of inadequate DNAse treatment or a high background splicing inefficiency resulting in a steady quantity of intron-mapped reads against fluctuating exon-mapped reads. This would appear as if there was a change in splicing ratio but it would meaningless.
However, there was a strong correlation between alternatively mapped reads and exonic reads, suggesting they scale together and therefore the relationship is not an illusion caused by static alternative read mapping.
Why this occurs is curious. Studies have shown there is a negative correlation between alternative splicing and gene expression under biotic stress (NMD-mediated regulation of plant gene expression and unspliced transcript sequestration in nuclear speckles are both possiblities.
